Instance segmentation
-
CartoCell
This tutorial describes how to train and infer using our custom ResU-Net 3D DNN in order to reproduce the results obtained in
(Andrés-San Román, 2022). Given an initial training dataset of 21 segmented epithelial 3D cysts acquired after confocal microscopy, we follow the CartoCell pipeline (figure below) to high-throughput segment hundreds of cysts at low resolution automatically.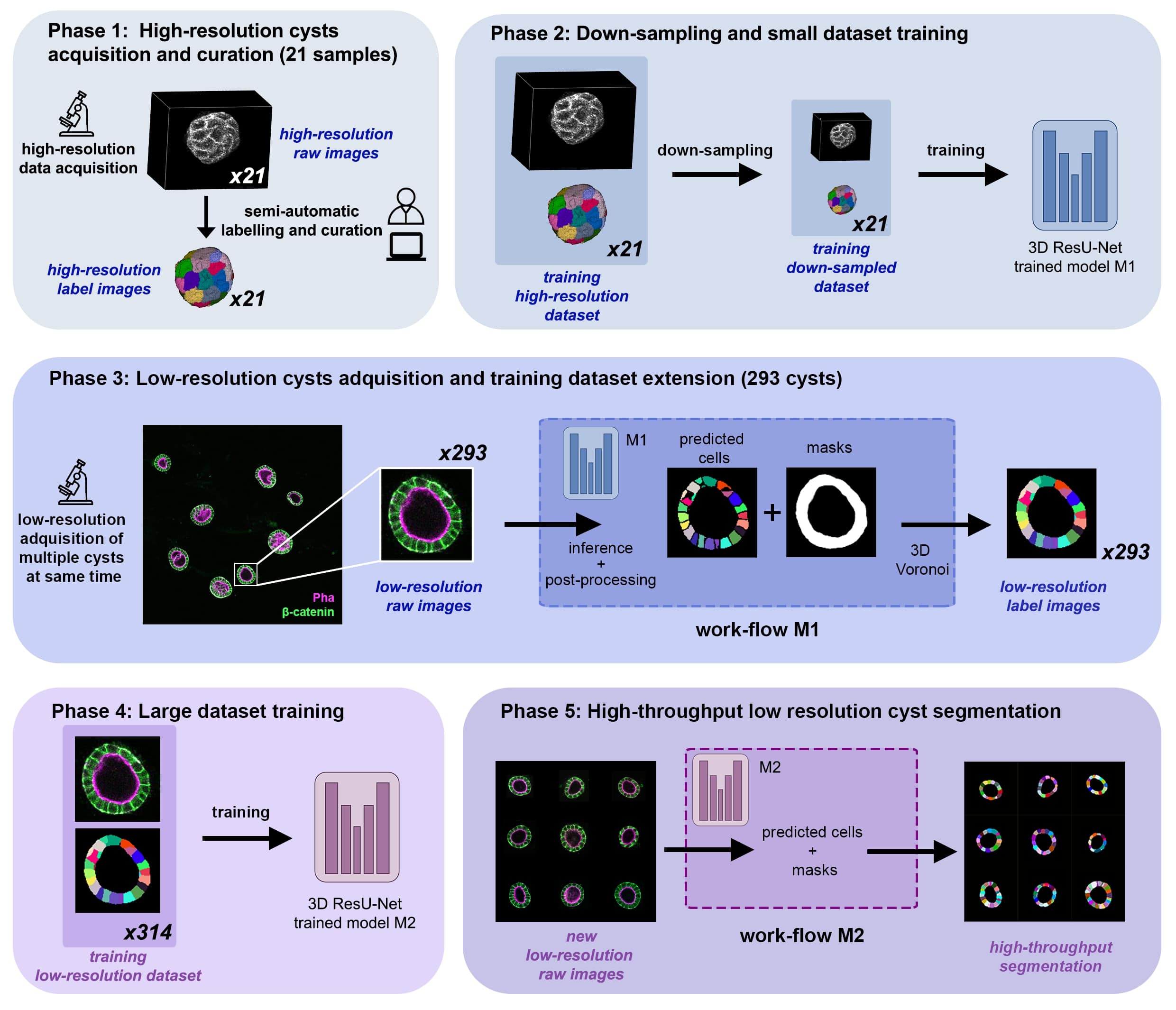
Citation:
Andres-San Roman, Jesus A., et al. "CartoCell, a high-content pipeline for 3D image analysis, unveils cell morphology patterns in epithelia." Cell Reports Methods 3.10 (2023). -
MitoEM dataset
The goal is to segment and identify automatically mitochondria instances in EM images. To solve such task pairs of EM images and their corresponding instance segmentation labels are provided. Below a pair example is depicted:
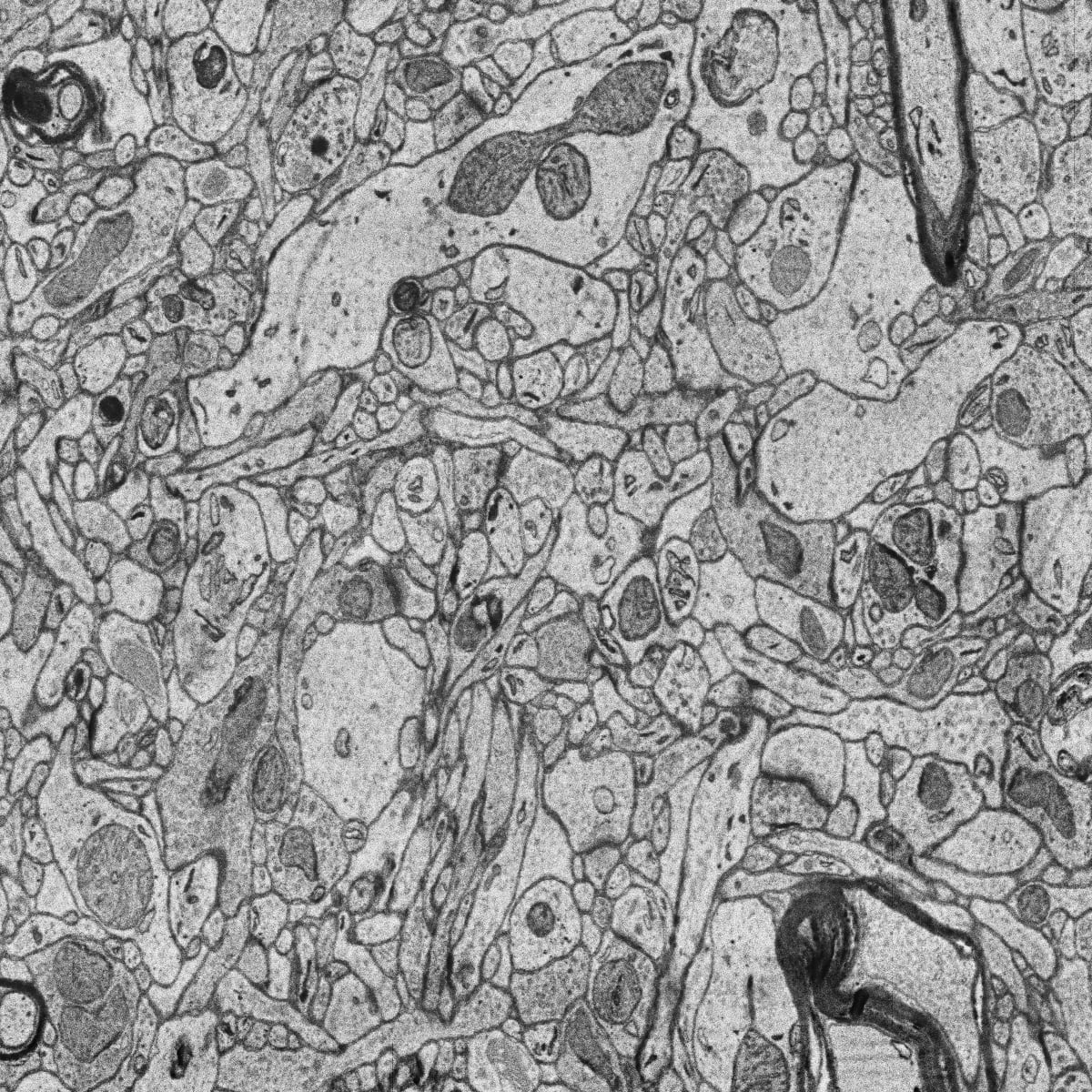

MitoEM dataset is composed by two EM volumes from human and rat cortices, named MitoEM-H and MitoEM-R respectively. Each volume has a size of
(1000,4096,4096)voxels, for(z,x,y)axes. Both tissues contain multiple instances entangling with each other with unclear boundaries and complex morphology, e.g., (a) mitochondria-on-a-string (MOAS) instances are connected by thin microtubules, and (b) multiple instances can entangle with each other.Citation:
Wei, Donglai, et al. "Mitoem dataset: Large-scale 3d mitochondria instance segmentation from em images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer International Publishing, 2020.
Semantic segmentation
-
Stable deep neural network architectures
The goal is to segment automatically mitochondria in EM images as described in Semantic segmentation. This is a semantic segmentation problem where pairs of EM image and its corresponding mitochodria mask are provided. Our purpose is to segment automatically other mitochondria in images not used during train labeling each pixel with the corresponding class: background or foreground (mitochondria in this case). As an example, belown are shown two images from EPFL Hippocampus dataset used in this work:
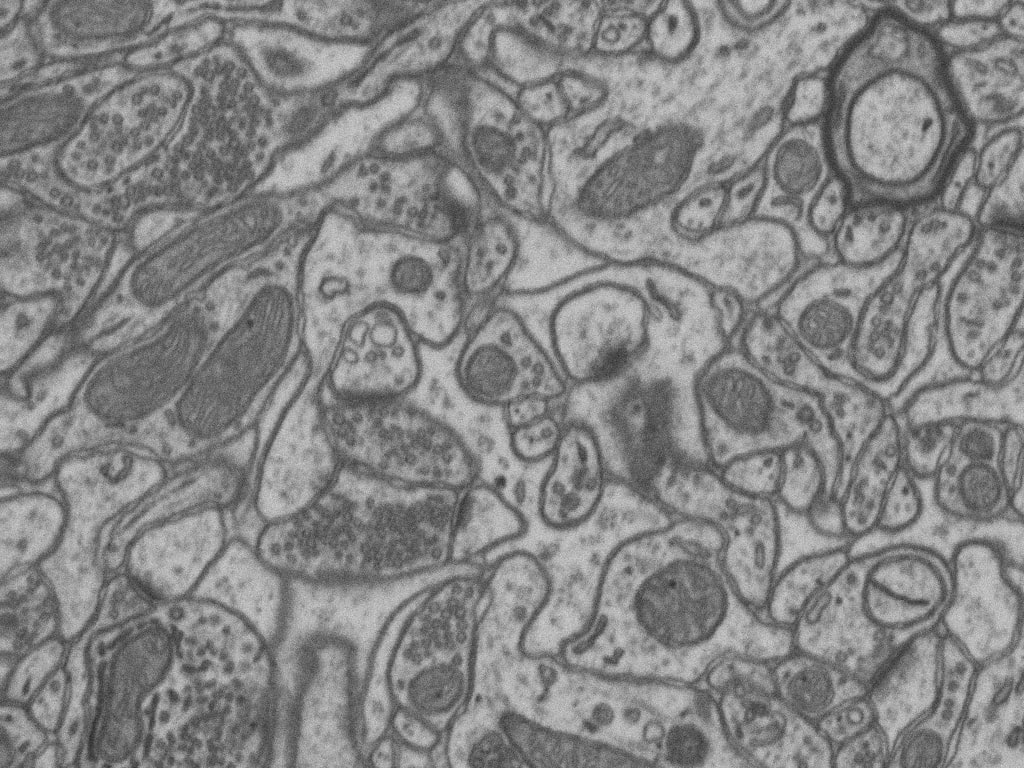

Citation:
Franco-Barranco, Daniel, Arrate Muñoz-Barrutia, and Ignacio Arganda-Carreras. "Stable deep neural network architectures for mitochondria segmentation on electron microscopy volumes." Neuroinformatics 20.2 (2022): 437-450.